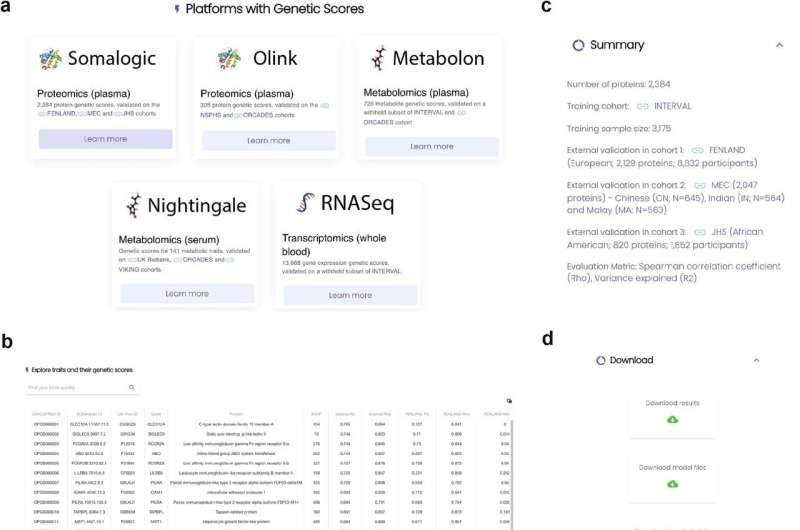
Work by an international research team led by Yu Xu and Michael Inouye at the Department of Public Health and Primary Care, University of Cambridge, has resulted in a unique resource for predicting multi-omics data directly from genotypes. The details of how the team developed the OmicsPred resource are in an article, "An atlas of genetic scores to predict multi-omic traits," published in the journal Nature. A Research Briefing on the study is published in the same journal issue.
There is a vision of the future that includes a device like the medical tricorder in "Star Trek," a hand-held device or an app incorporated into a mobile device. After a quick and painless scan just by waving the device over the afflicted area, comes the prognosis—such as Barclay's protomorphosis syndrome, a pernicious DNA disease that makes one de-evolve in the most intriguing science-fiction way possible. While a device of this type remains science fiction, many of the medical tricorders' capabilities already exist in the form of bulky lab equipment and databases spread across the fields of omics.
Thorough investigation of disease, or disease susceptibility, requires a lot of different omics—genomics, epigenomics, transcriptomics, proteomics and metabolomics. The collection of multi-omics is costly and data-intensive, making it somewhat rare within research. These fields of knowledge are in databases full of detailed analyses of human cell functions and disease associations. Additionally, many multi-omics studies have been targeted at specific subset populations to interrogate disease mechanisms. A broader capture of multi-omics could confirm inferred knowledge and discover hidden biological pathways.
Xu and colleagues used a machine-learning approach to create genetic scores for 17,227 biomolecular traits from 48,813 healthy blood samples that can predict the levels of 13,668 RNA transcripts, 2,692 proteins and 867 metabolites. The genetic scores were then validated in seven diverse, independent cohorts.
The OmicsPred team anticipates that this new resource will be widely useful in investigating multi-omic traits and associations with biological traits. All the genetic scores from the study have been made public through the web portal omicspred.org, and the scientific community is invited to use it to predict multi-omic traits from genotypes in their own data sets.
Currently, the molecular traits that these scores predict only reflect the heritability and variability in the training data set, which was derived from healthy blood donors of predominantly white European ancestries. The creators plan to enhance and refine the range of genetic scores available in the OmicsPred resource and expand the ancestral diversity with updated training data sets.
© 2023 Science X Network








 User Center
User Center My Training Class
My Training Class Feedback
Feedback












Comments
Something to say?
Login or Sign up for free